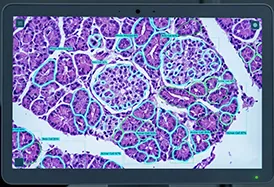
 A global leader in therapeutic antibody discovery faced a major engineering bottleneck in their high-content screening (HCS) workflow. Their microscopy hardware was generating terabytes of single-cell imagery, but their manual analysis processes couldn’t keep up. They needed a partner to engineer a scalable, automated solution. By partnering with Cardinal Peak / FPT for Custom Bioimage Analysis Software Development, they deployed an end-to-end computer vision pipeline. Our solution automated the instance segmentation and classification of millions of B-cells, increasing throughput by 10x and improving usable cell identification by 30%.
A global leader in therapeutic antibody discovery faced a major engineering bottleneck in their high-content screening (HCS) workflow. Their microscopy hardware was generating terabytes of single-cell imagery, but their manual analysis processes couldn’t keep up. They needed a partner to engineer a scalable, automated solution. By partnering with Cardinal Peak / FPT for Custom Bioimage Analysis Software Development, they deployed an end-to-end computer vision pipeline. Our solution automated the instance segmentation and classification of millions of B-cells, increasing throughput by 10x and improving usable cell identification by 30%.
The Challenge: Engineering for Terabyte-Scale Image Data
The client’s platform uses transgenic animal models to generate massive libraries of antibodies. The engineering challenge was building a system capable of identifying rare, high-value B-cells hidden within millions of images generated by automated microscopy.
Their existing infrastructure and off-the-shelf software failed at scale:
- Data Deluge: A single screening run generated terabytes of high-resolution images. Local servers and standard software choked on the volume, creating a massive backlog.
- Segmentation Failure: Standard image analysis tools using simple thresholding could not reliably perform instance segmentation on clustered or overlapping cells, leading to inaccurate data.
- Latency: The time lag in processing meant biological samples often degraded before “hits” could be identified and isolated by robotic systems. They required a custom-engineered, cloud-native AI pipeline capable of real-time processing.
Building a pipeline for terabyte-scale image data isn’t just about AI; it’s a massive data engineering challenge. We moved the workflow to a cloud-native architecture to eliminate on-premise bottlenecks, allowing millions of cells to be segmented and classified in real-time.
The Engineering Solution: An End-to-End Deep Learning Pipeline
We engineered a custom high-throughput screening AI pipeline, leveraging our partnership with Landing AI to deploy robust computer vision models at scale.
High-Throughput Data Engineering
We moved the workflow off-premise, architecting a cloud-native data pipeline. Images stream directly from microscopy instruments to scalable cloud storage. This triggers automated pre-processing steps (normalization, artifact removal) and queues images for parallelized inference, eliminating the on-premise bottleneck.
Deep Learning for Instance Segmentation
To solve the clustering problem, we abandoned traditional vision techniques in favor of custom deep learning models trained for instance segmentation. The AI accurately delineates each individual B-cell as a unique object, even in dense clusters, ensuring precise measurements for every single candidate.
Automated Classification & Robot Integration
Following segmentation, a second model classifies each cell based on multi-channel fluorescent signals, scoring them for therapeutic potential based on expert-annotated training data. The system generates a structured output file of “pick coordinates,” integrating directly with downstream liquid handling robots for automated isolation.
High-Content Screening AI Pipeline Architecture

The Results: 10x Throughput & Higher Yield
The engineered pipeline transformed a manual R&D bottleneck into a scalable, automated production workflow.
- 10x Increase in Throughput: Automated analysis reduced screening timelines from weeks to days, allowing the client to process significantly larger antibody libraries.
- 30% Improvement in Usable Cell Identification: More accurate segmentation and classification led to a higher yield of viable, high-value B-cells for downstream development.
- Scalable Infrastructure: The cloud-native architecture decoupled processing from local hardware, allowing infinite scalability as data volumes grow.
Core AI
- Deep Learning (CNNs)
- Instance Segmentation
Platform
- i2 Vision / Landing AI partnership for visual inspection
Infrastructure
- Cloud-native data pipeline
- GPU-accelerated compute
The Engine Behind the Solution
This project was accelerated by our AI Center of Excellence (CoE). By leveraging pre-trained computer vision models and enterprise-grade MLOps infrastructure, we delivered a scalable, high-performance solution tailored to the rigorous demands of biopharma R&D.
Our AI Center of Excellence Capabilities
Explore Engineering Services Related to Life Sciences AI

Custom Computer Vision Development
Discover our broader capabilities in analyzing complex images and video across industries.
Bioimage Analysis Software Development FAQs
Why is custom software development necessary for high-content screening (HCS)?
Off-the-shelf bioimage software often fails at the scale and complexity of modern pharma R&D. Custom bioimage analysis software development is required to build pipelines that can handle terabyte-scale datasets, integrate with specific legacy laboratory hardware (like microscopes and robots), and utilize specialized deep learning models trained on a company’s unique biological data.
What is "instance segmentation" and why is it critical for single-cell analysis?
Semantic segmentation just labels pixels as “cell” or “background.” Instance segmentation goes a step further, identifying each individual cell as a unique, discrete object, even if they are touching in a cluster. This is absolutely critical for B-cell screening, as you must measure the specific biological activity of single, distinct candidates to select the right ones for cloning.
How do engineering teams handle terabyte-scale microscopy data?
Handling terabytes of daily image data requires moving away from local servers to cloud-native engineering. We architect solutions that stream images directly from instruments to scalable cloud object storage (like S3), using event-driven architecture to trigger parallel GPU compute clusters for rapid analysis.
How does deep learning improve upon traditional image analysis in biology?
Traditional analysis relies on rigid manual rule-setting (e.g., “count anything brighter than X”). These rules fail when faced with biological variability and background noise. Deep learning models learn features directly from expert-annotated data, allowing them to robustly handle noisy images, clustered cells, and subtle variations just as a trained biologist would, but at massive scale.
Can these computer vision pipelines integrate with laboratory robotics?
Yes. A key part of the engineering effort is the final integration layer. The AI pipeline doesn’t just produce a picture; it outputs structured data—such as precise well coordinates of identified “hits”—in formats that can be directly ingested by automated liquid handling and cell-picking robots, closing the loop on automation.





